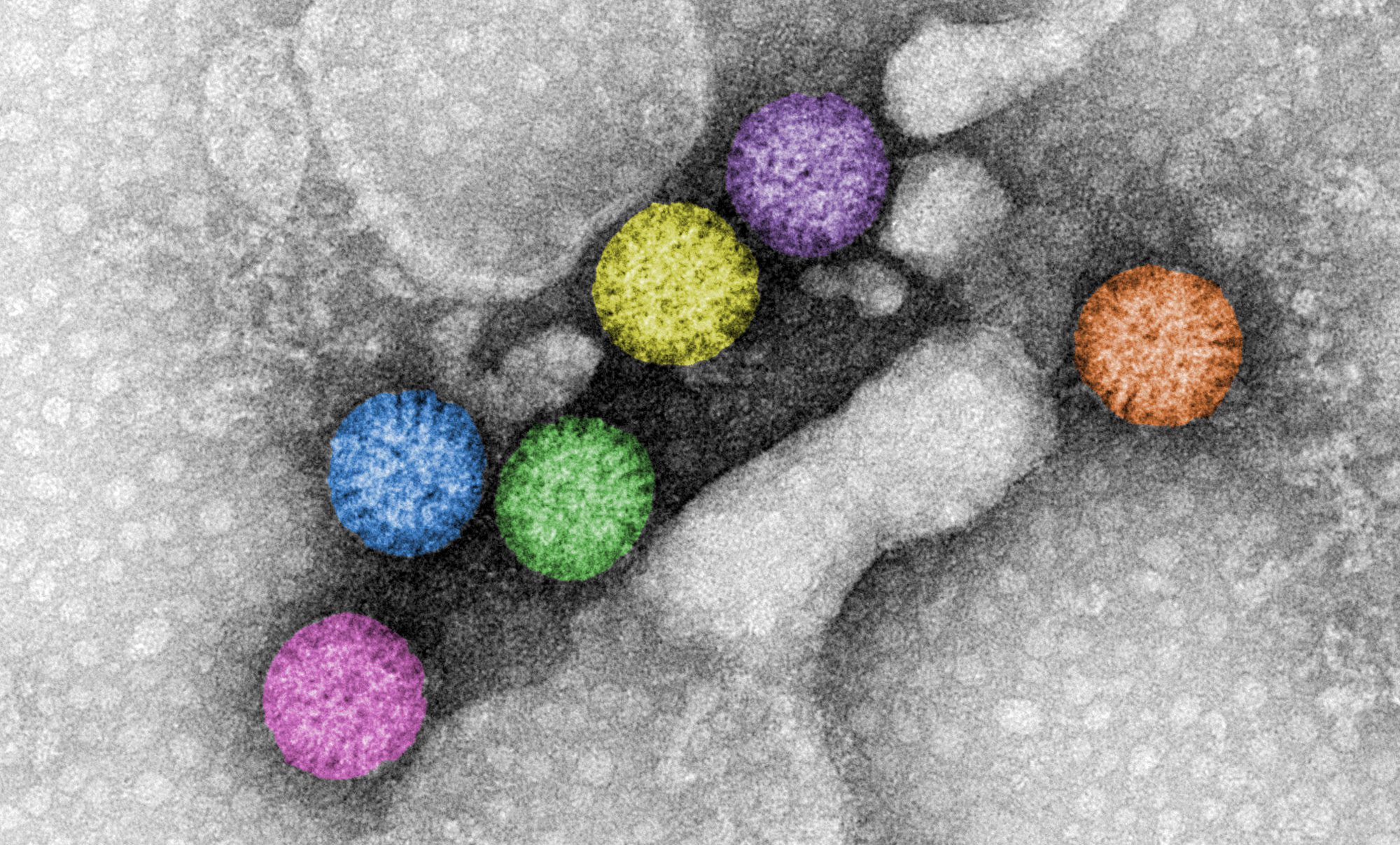
Rotavirus triple-layered particles. Rotavirus virions are triple-layered particles with icosahedral symmetry. The spike protein and the glycoprotein VP7 compose the external layer. Meanwhile, a unique component, VP6, is part of the middle layer. The VP2 icosahedral internal layer or virus core encapsidates the eleven dsRNA genome segments. All twelve vertices of inner core contain VP1-VP3 complexes. The image represents high-definition transmission electron microscopy of rotavirus triple-layered particles purified from cell culture. After glutaraldehyde fixation, the particles were stained phosphotungstate. Acquired images were colored using Adobe Photoshop.
Courtesy of Catherine Eichwald, Institute of Virology, University of Zürich.
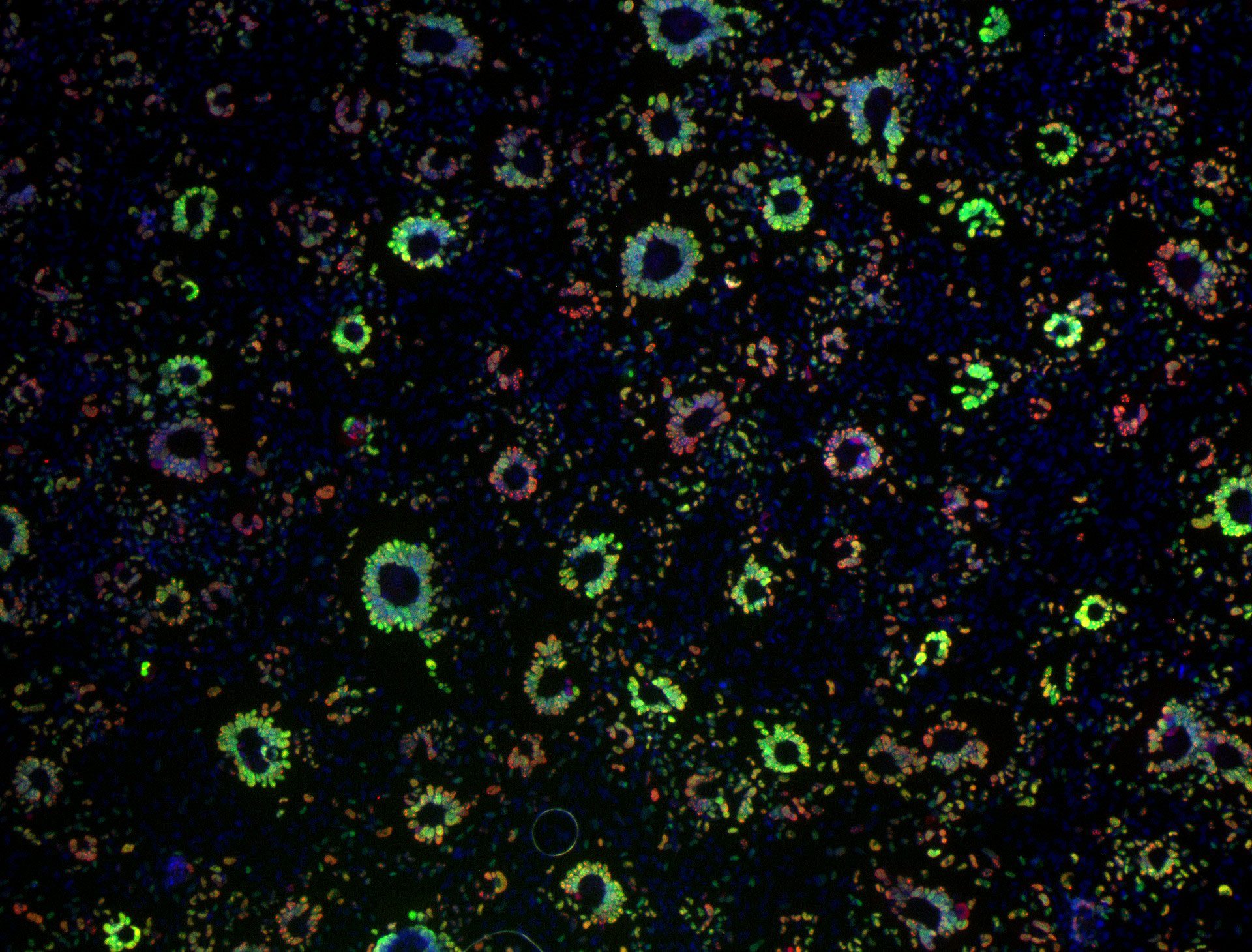
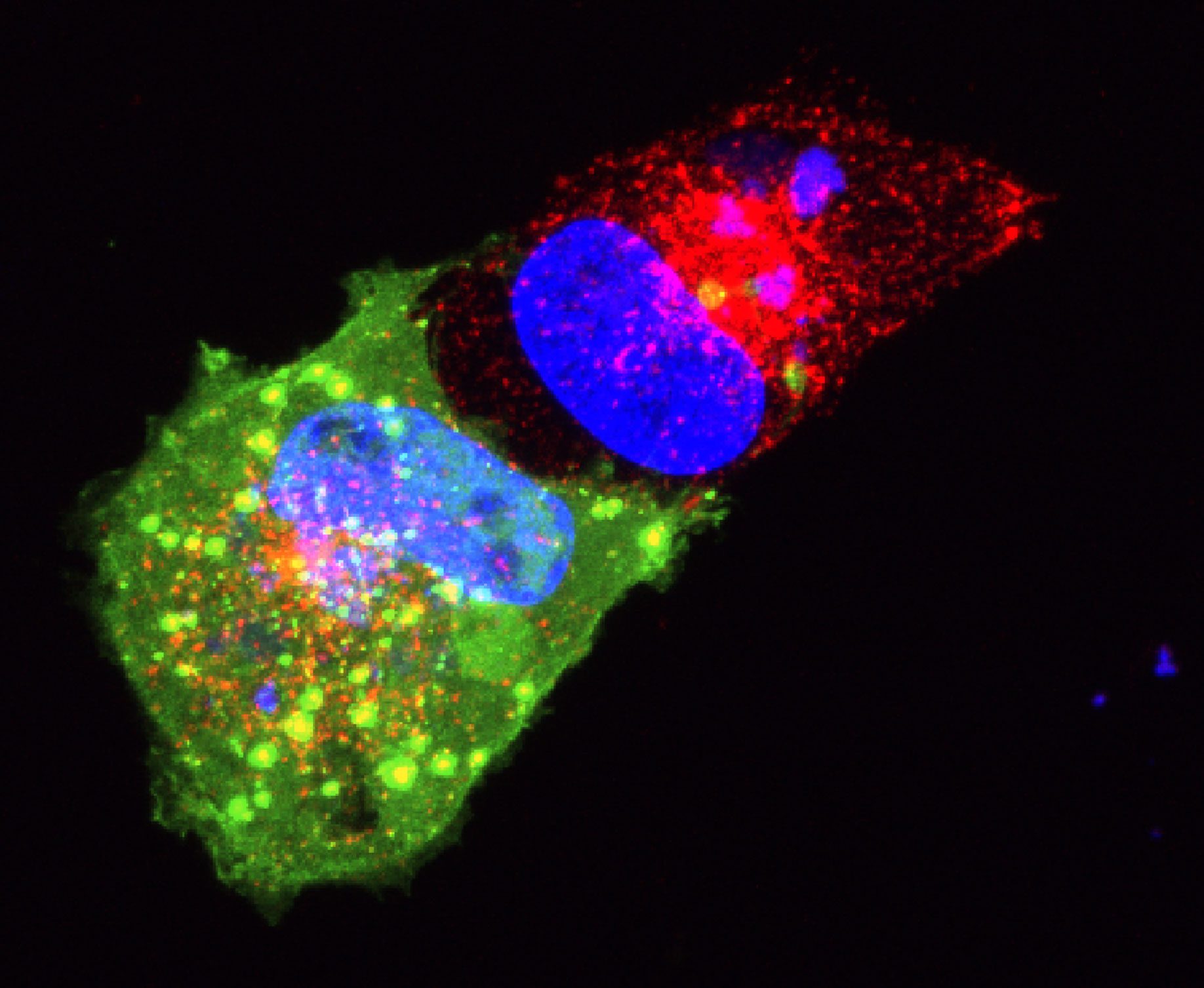
Epithelial cell monolayer (ARPE-19) infected with human cytomegalovirus, costained for viral immediate-early proteins 1/2 (green) and viral single-stranded DNA binding protein UL57 (red). Nuclei are stained with Hoechst 33342 (blue). See Vo, M., A. Aguiar, M.A. McVoy, and L. Hertel. 2020. Cytomegalovirus Strain TB40/E Restrictions and Adaptations to Growth in ARPE-19 Epithelial Cells. Microorganisms 8:E615. doi: 10.3390/microorganisms8040615.
Courtesy of Alexis Aguiar and Laura Hertel, UC San Francisco.
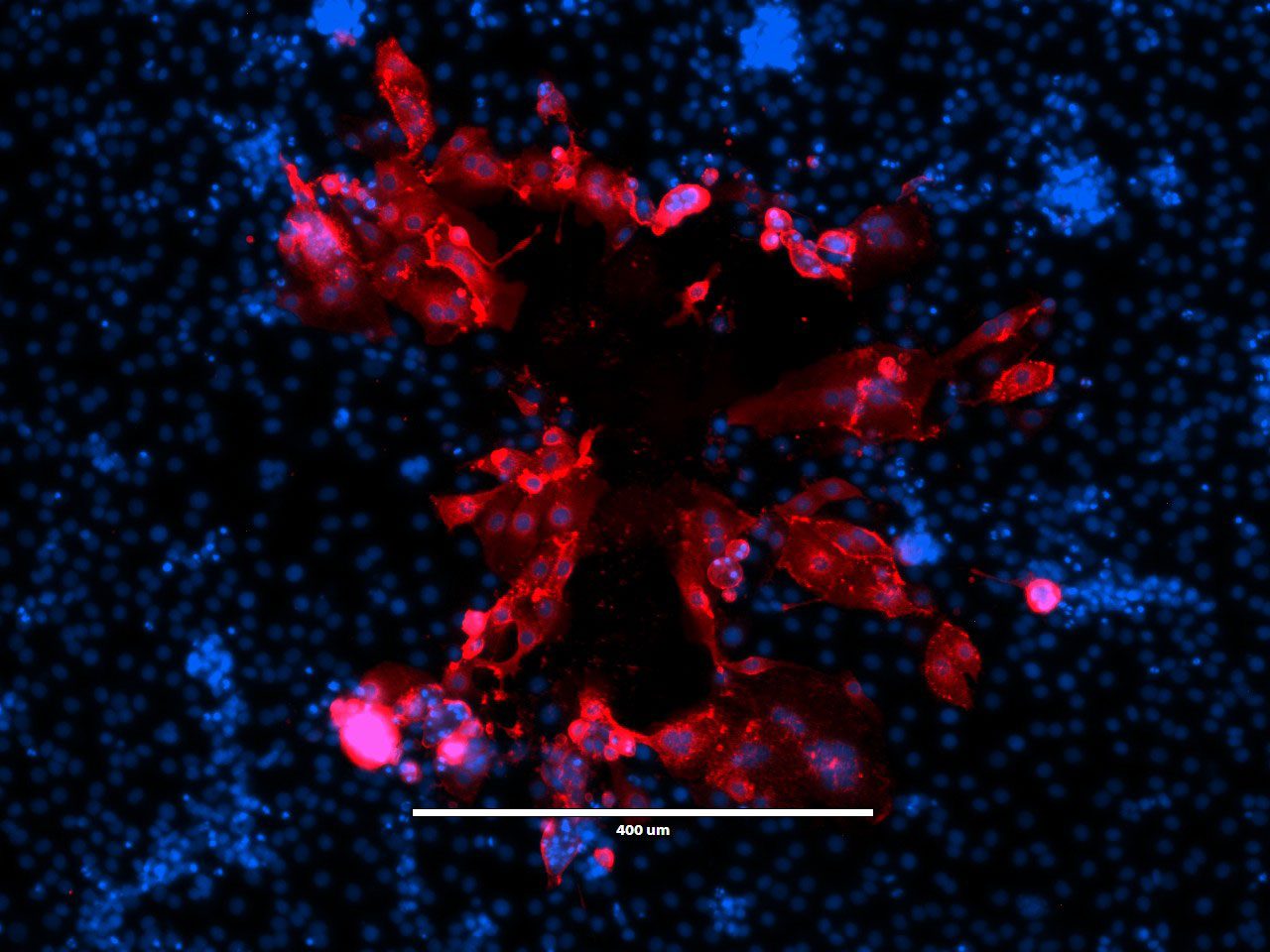
“Beauty Is Infectious.” Immunofluorescence image of a virus plaque formed by human metapneumovirus (HMPV) in LLC-MK2 cells. HMPV (red) and nucleus (DAPI, blue).
Image courtesy of Jiuyang Xu (visting scholar, Tsinghua University School of Medicine) and John V. Williams, Department of Pediatrics, University of Pittsburgh.
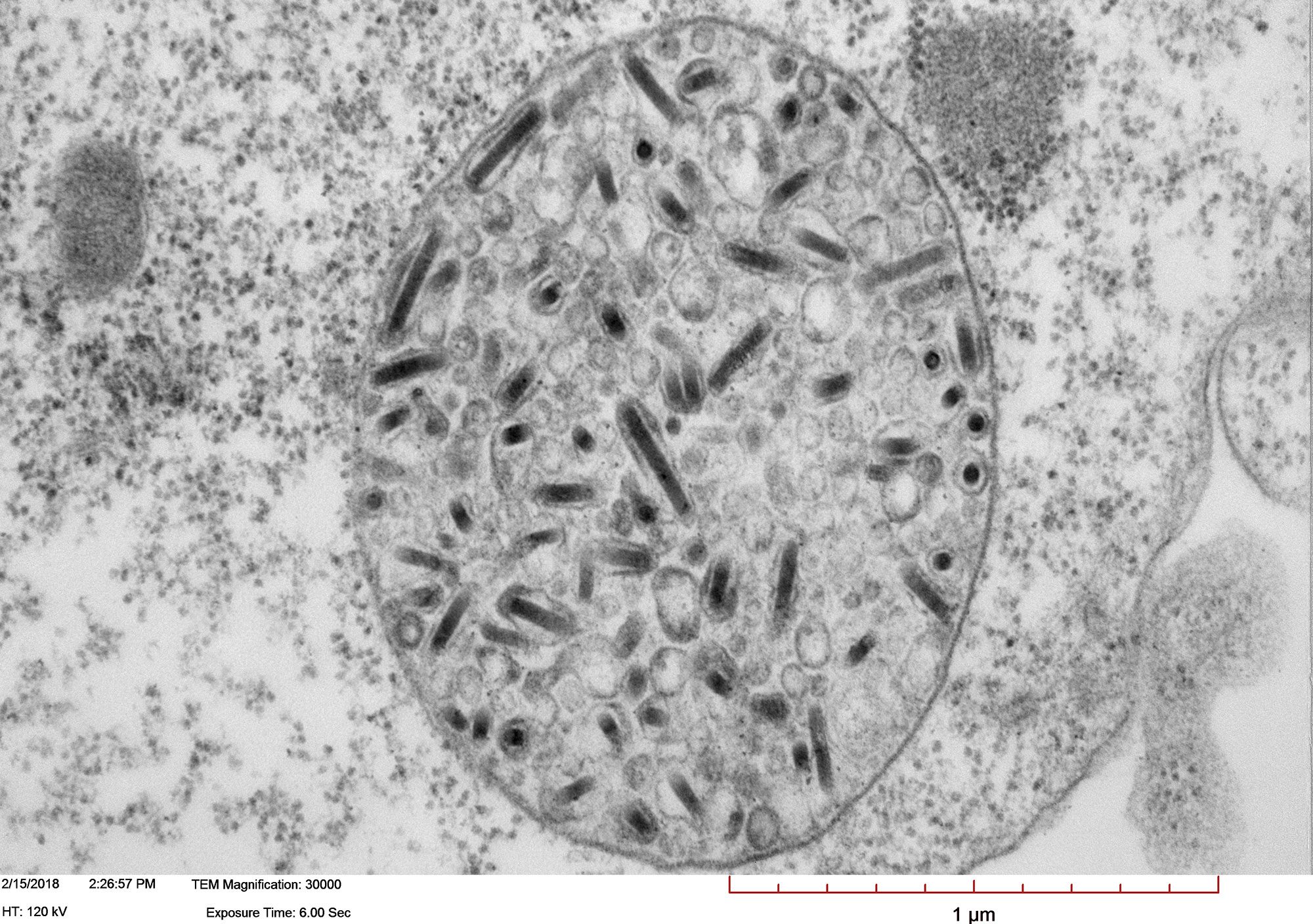
Transmission electron micrograph of High Five cells infected with Autographa californica multiple nucleopolyhedrovirus (AcMNPV). Image shows a large vesicle containing more than one enveloped nucleocapsid.
Courtesy of Xiao-Wen Cheng, Miami University.
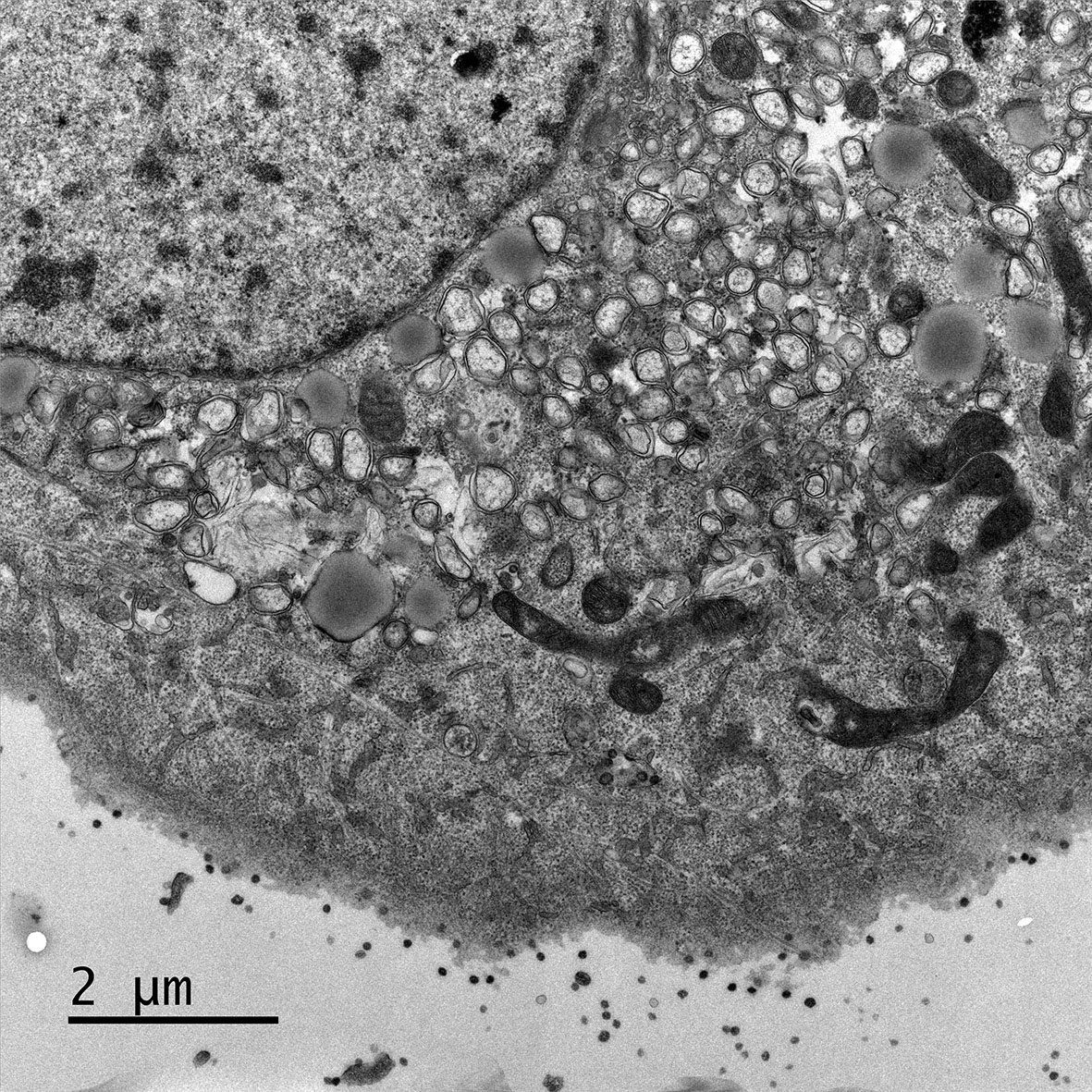
Electron micrograph of Vero E6 cells infected with SARS-CoV-2, 8 h post infection. Viral replication organelles. These virus-induced structures accumulate in large clusters in the perinuclear region and primarily contain double-membrane vesicles (DMVs). See Ogando et al., “SARS-Coronavirus-2 Replication in Vero E6 Cells: Replication Kinetics, Rapid Adaptation, and Cytopathology.” 2020. J. Gen. Virol. doi: 10.1099/jgv.0.001453 PMID 32568027.
Courtesy of Ronald Limpens, Montse Bárcena and Eric Snijder, Leiden University Medical Center, the Netherlands.
SVG-A glial cells transiently transfected with dominant negative dynamin (green) treated with transferrin-594 (red), and stained with DAPI (blue). 5 minutes post internalization.
Courtesy of Colleen Mayberry and Melissa Maginnis, University of Maine.

Well-differentiated small airway epithelial cells infected with recombinant human respiratory syncytial virus expressing enhanced green fluorescent protein at 3 days post-infection. Green, recombinant human respiratory syncytial virus; orange, acetylated α-tubulin (cilia) ; magenta, zona occludens 1 (tight junctions). 40x magnification.
Courtesy of Laurine Rijsbergen, Department of Viroscience, Erasmus Medical Center, Rotterdam, The Netherlands.

SVG-A glial cells infected with JC polyomavirus labeled with Alexa Fluor 647 (pseudocolored magenta), 5 minutes post internalization. Cells are stained for actin (pseudocolored gray); nuclei are stained with DAPI (blue).
Courtesy of Colleen Mayberry and Melissa Maginnis, University of Maine.
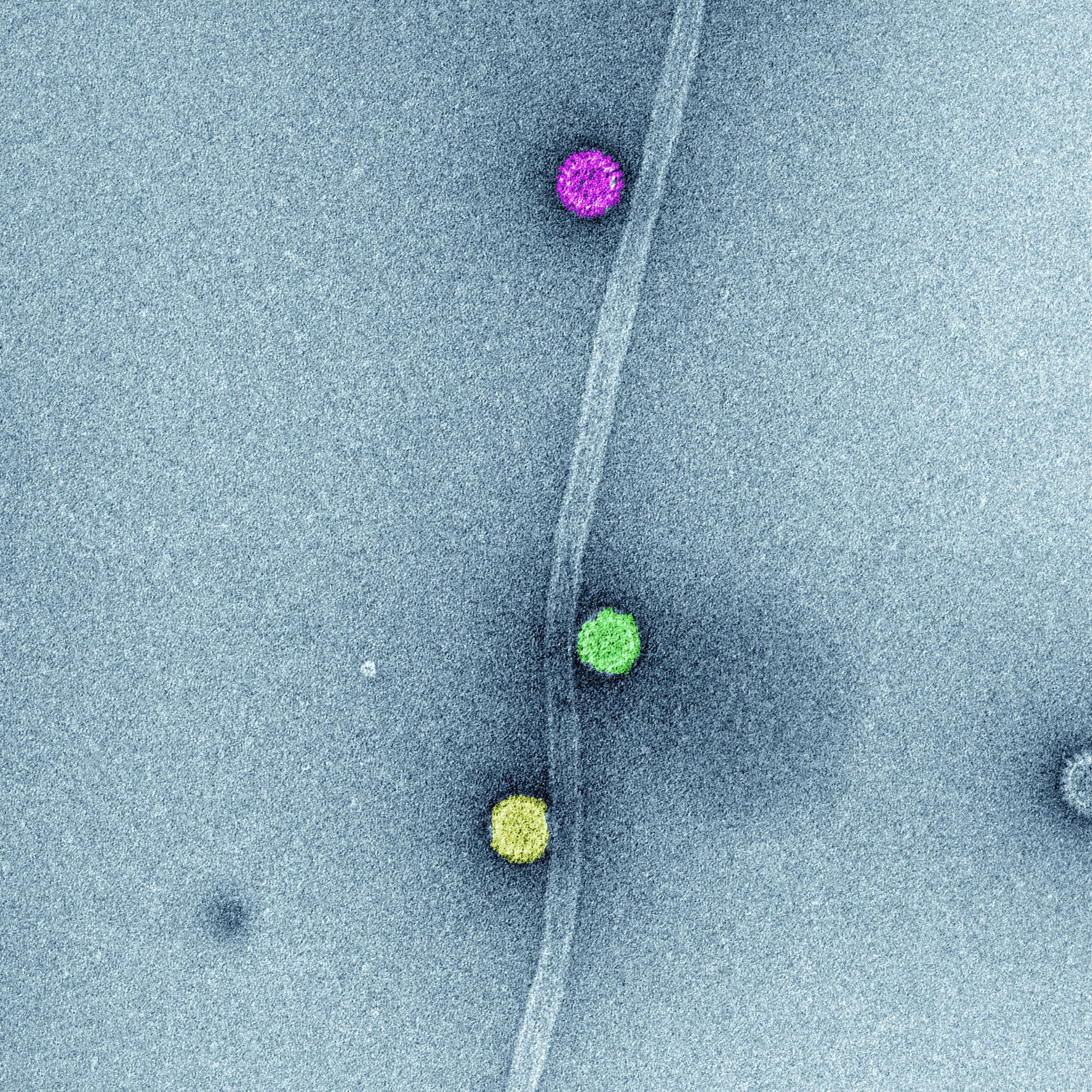
In vitro association of rotavirus triple-layered particle to microtubules (MT). In vitro synthesized microtubules and purified rotavirus triple-layered particles were spinned-down in a sucrose cushion. Samples of resuspended pellets were analyzed by high definition transmission electron microscopy to determine the direct association among rotavirus triple-layered particles and MTs. The image represents high-definition transmission electron microscopy of sedimented MTs and rotavirus triple-layered particles. After glutaraldehyde fixation, the sample was stained with phosphotungstate. Acquired images were colored using Adobe Photoshop.
Courtesy of Catherine Eichwald, PhD, Institute of Virology, University of Zürich
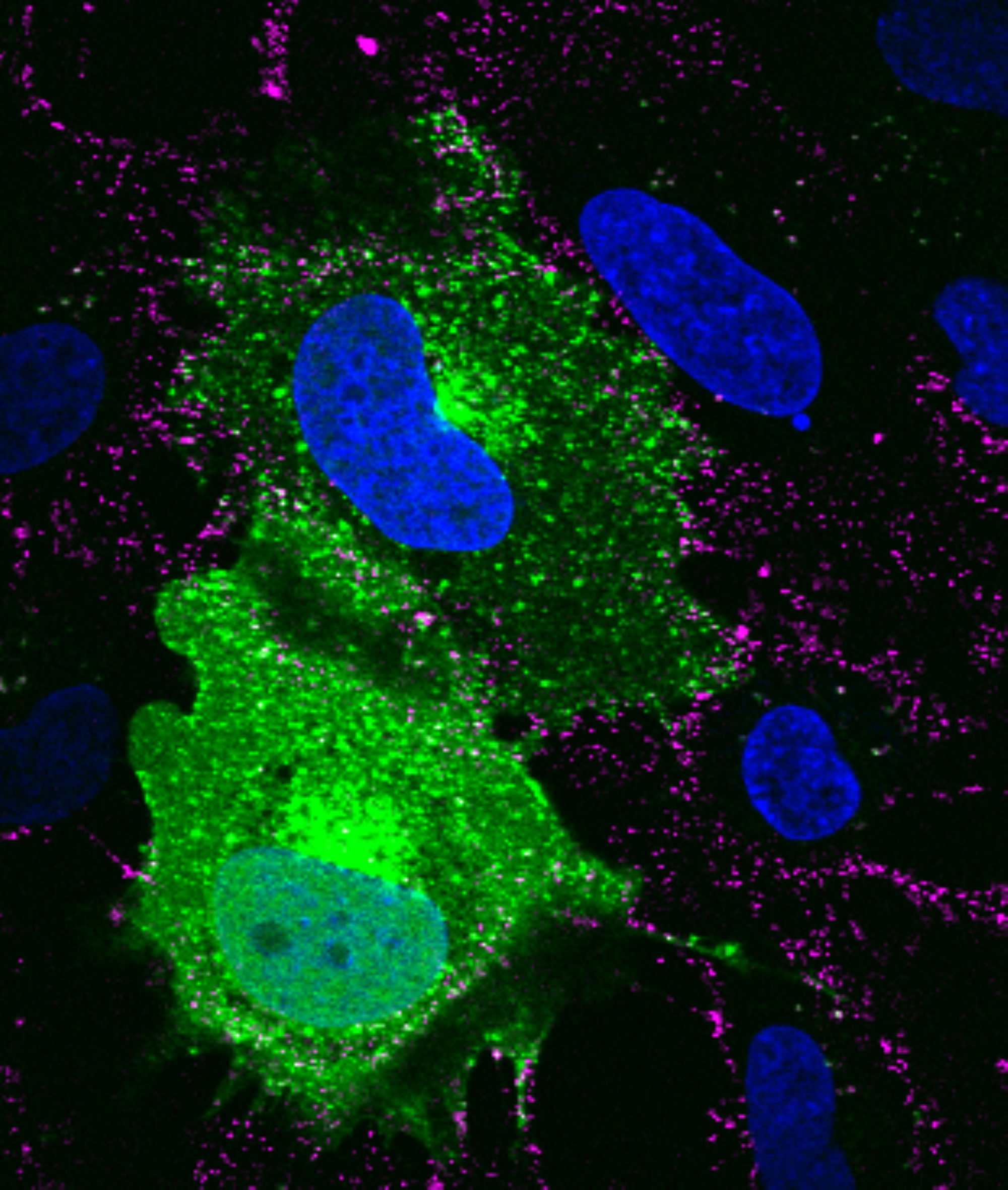
SVG-A glial cells transiently transfected with mEmerald clathrin (green), infected with JC polyomavirus labeled with Alexa Fluor 647 (pseudocolored magenta), 5 minutes post internalization. Nuclei are stained with DAPI (blue).
Courtesy of Colleen Mayberry and Melissa Maginnis, University of Maine.
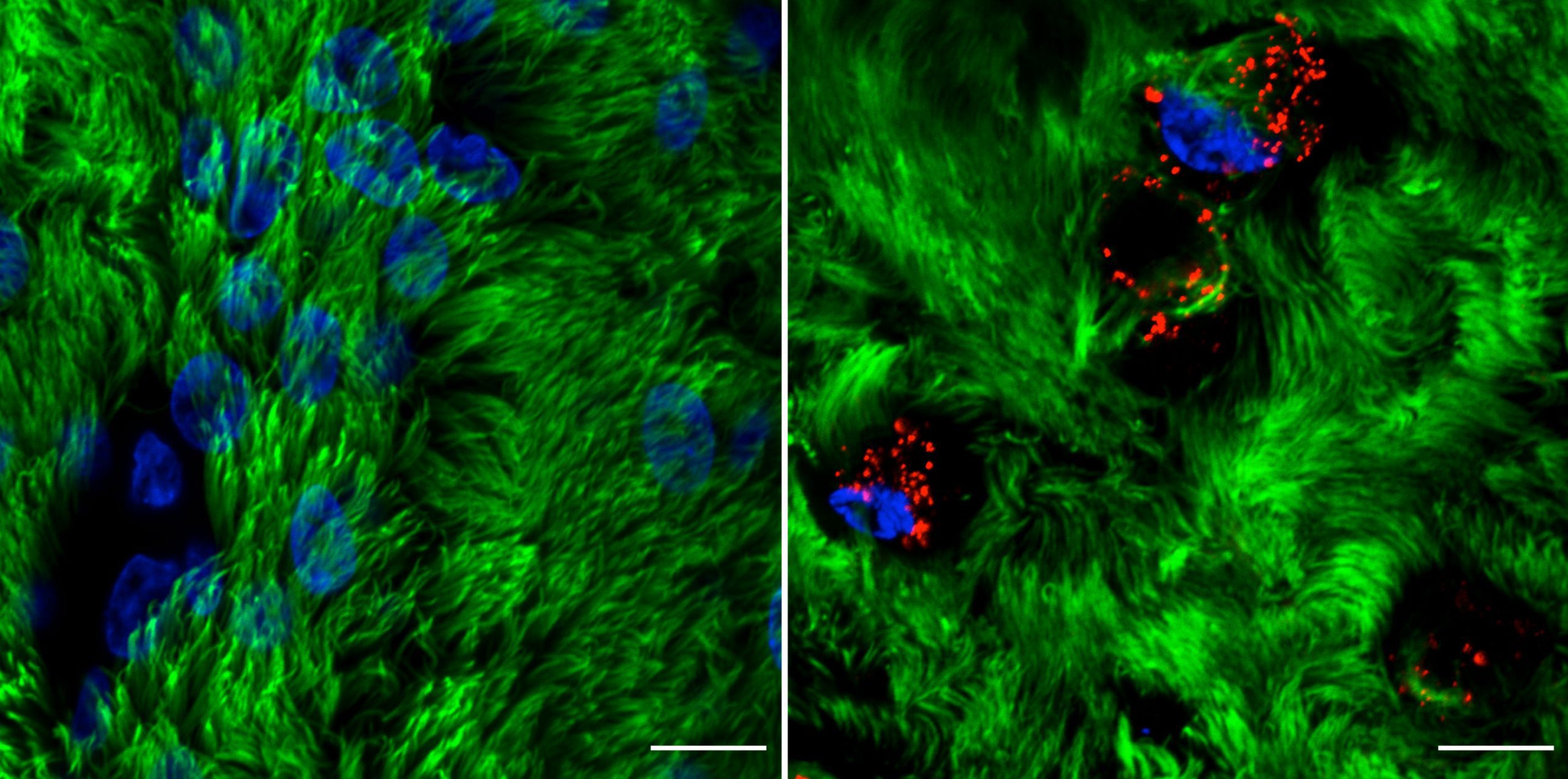
Rhinovirus C15 (RV-C15) infection of well-differentiated human bronchial epithelial cells cultured in air-liquid interface (hBEC-ALI) fixed at 12h post-infection (p.i.). A total of 50 μL of inoculum (PBS, left; or 1012 copies of RNA of RV-C15, right) was added onto the top of hBEC and incubated at 34°C. The inoculum was removed 4 h p.i., and the cells were fixed at 12h p.i. with 4% paraformaldehyde. The indirect immunofluorescence assay was done using antibody against acetylated alpha-tubulin to detect motile cilia (#ab24610, Abcam; green-colored) and against dsRNA to detect the replicative intermediary forms of RV-C15 (#10010200, Scicons; red-colored); while the nuclei were stained with Hoechst 33342 (#H3570, Molecular Probes; blue-colored). The images were acquired using confocal microscopy (LSM710, Carl Zeiss Microscopy) at 630x magnification (scale bar = 10 μM).
Courtesy of Talita Gagliardi, University of Maryland, College Park.
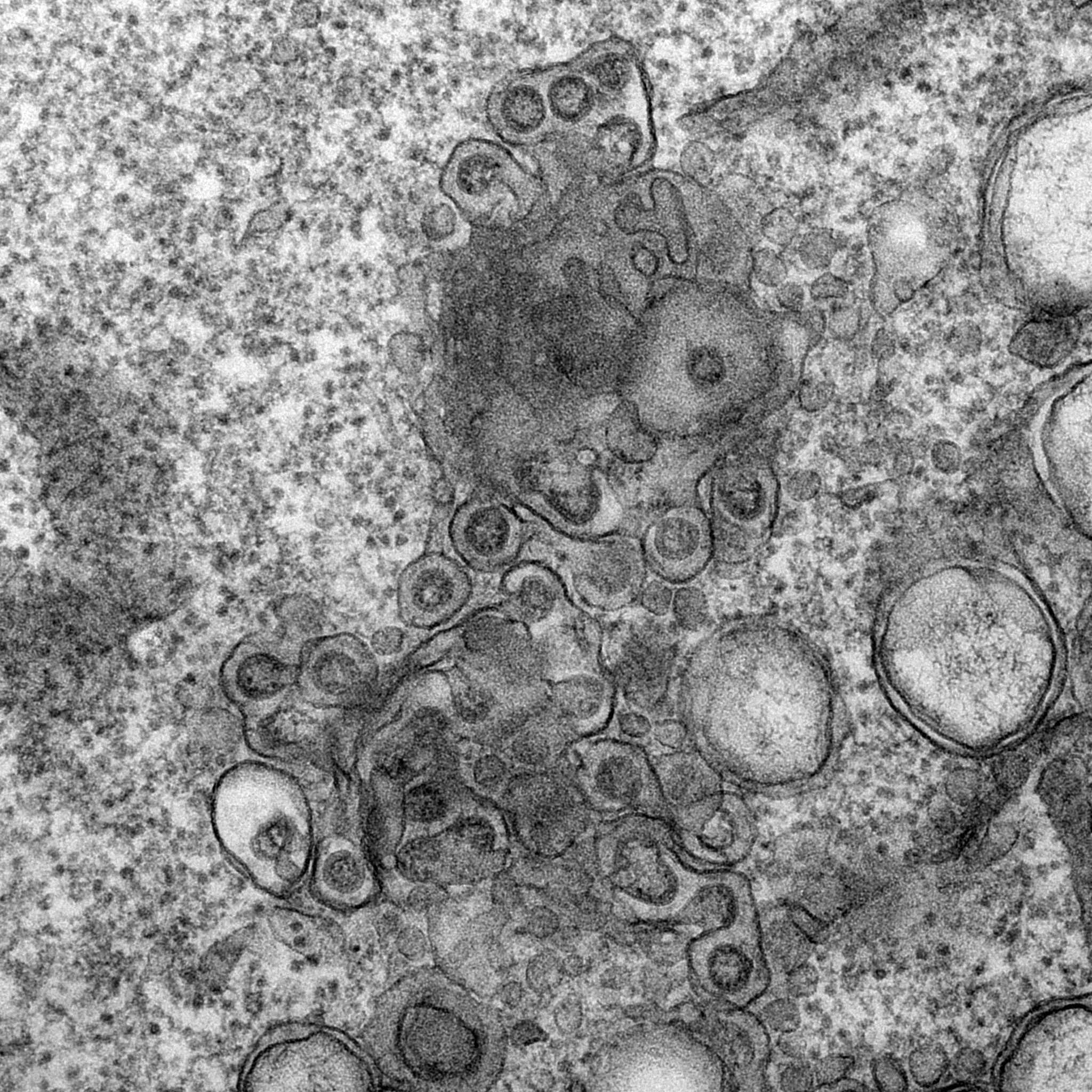
Electron micrograph of Vero E6 cells infected with SARS-CoV-2, 10 h post infection. Virus particles budding into membranes of the ERGIC. See Ogando et al., “SARS-Coronavirus-2 Replication in Vero E6 Cells: Replication Kinetics, Rapid Adaptation, and Cytopathology.” 2020. J. Gen. Virol. doi: 10.1099/jgv.0.001453 PMID 32568027.
Courtesy of Ronald Limpens, Montse Bárcena and Eric Snijder, Leiden University Medical Center, the Netherlands.
