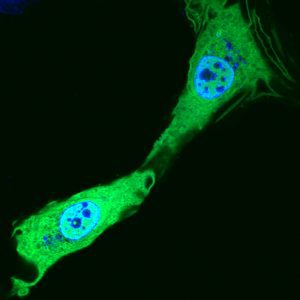
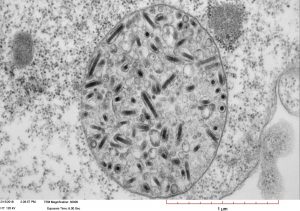
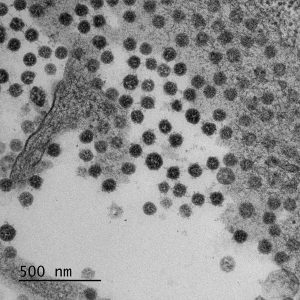
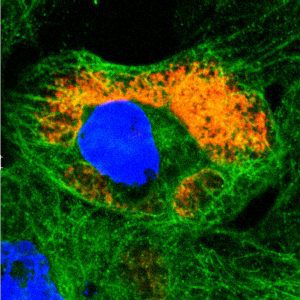
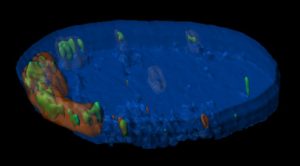
Saccharomyces cerevisiae infected with double-stranded RNA totiviruses and associated satellites enable the production of antifungal killer toxins. Courtesy of Paul Rowley, Department of Biological Sciences, University of Idaho.
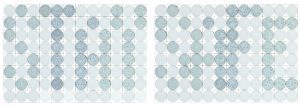
“STAY SAFE.” Wells spelling this important expression during the COVID-19 pandemic were created using a focus-forming assay for SARS-CoV-2 and Vero E6 cells. In each plate, columns 1-4 were seeded with 3 x 104 cells/well; columns 5-8, 1.5 x 105 cells/well; columns 9-12, 3 x 105 cells/well. After one day, virus was added to some wells at each density and incubated 24 h at 37°C prior to fixation and development by incubation with a polyclonal antibody to detect SARS-CoV-2 spike protein, followed by a secondary HRP-labeled antibody. A p1 SARS-CoV-2 stock grown in Huh7.5 cells was used for the inoculations. In the images, wells where no virus was added were lightened so the letters (foci-containing wells) would stand out. Courtesy of Amelia Pinto, James Brien, and E. Taylor Stone, Department of Molecular Microbiology and Immunology, Saint Louis University.
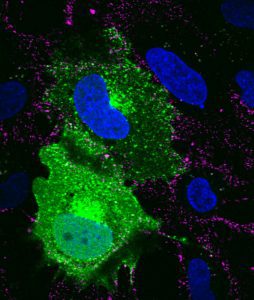
SVG-A glial cells transiently transfected with mEmerald clathrin (green), infected with JC polyomavirus labeled with Alexa Fluor 647 (pseudocolored magenta), 5 minutes post internalization. Nuclei are stained with DAPI (blue). Courtesy of Colleen Mayberry and Melissa Maginnis, University of Maine.
Figure 3. In vitro association of rotavirus triple-layered particle to microtubules (MT). In vitro synthesized microtubules and purified rotavirus triple-layered particles were spinned-down in a sucrose cushion. Samples of resuspended pellets were analyzed by high definition transmission electron microscopy to determine the direct association among rotavirus triple-layered particles and MTs. The image represents high-definition transmission electron microscopy of sedimented MTs and rotavirus triple-layered particles. After glutaraldehyde fixation, the sample was stained with phosphotungstate. Acquired images were colored using Adobe Photoshop.
SVG-A glial cells transiently transfected with mEmerald clathrin (green) prior to fixation. Nuclei are stained with DAPI (blue). Courtesy of Colleen Mayberry and Melissa Maginnis, University of Maine.
Rotavirus viroplasm. Rotavirus genome replication, packaging of virus genome segments in viral cores, and double-layered particle (DLP) formation occur in membrane- less cytosolic inclusions named viroplasms. Freshly made DLPs bud from viroplasms to rough endoplasmatic reticulum (RER) to become mature triple-layered particles (TLPs). In this process, a transient lipid membrane surrounds immature TLPs. The image represents a high-definition transmission electron micrograph of a viroplasm (magenta) at 6 h post-infection, surrounded by RER containing immature and mature TLPs (orange, yellow, green, red, light blue, dark blue). After glutaraldehyde fixation, samples were stained with uranyl acetate and lead citrate. Acquired images were colored using Adobe Photoshop. Courtesy of Catherine Eichwald, Institute of Virology, University of Zürich.
SVG-A glial cells infected with JC polyomavirus labeled with Alexa Fluor 647 (pseudocolored magenta), 5 minutes post internalization. Cells are stained for actin (pseudocolored gray); nuclei are stained with DAPI (blue). Courtesy of Colleen Mayberry and Melissa Maginnis, University of Maine.
Well-differentiated small airway epithelial cells infected with recombinant human respiratory syncytial virus expressing enhanced green fluorescent protein at 3 days post-infection. Green, recombinant human respiratory syncytial virus; orange, acetylated α-tubulin (cilia) ; magenta, zona occludens 1 (tight junctions). 40x magnification. Courtesy of Laurine Rijsbergen, Department of Viroscience, Erasmus Medical Center, Rotterdam, The Netherlands.
SVG-A glial cells transiently transfected with dominant negative dynamin (green) treated with transferrin-594 (red), and stained with DAPI (blue). 5 minutes post internalization. Courtesy of Colleen Mayberry and Melissa Maginnis, University of Maine.
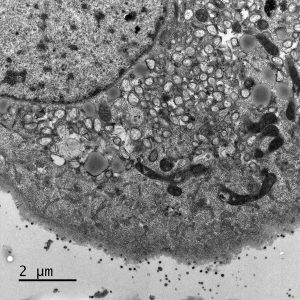
Electron micrograph of Vero E6 cells infected with SARS-CoV-2, 8 h post infection. Viral replication organelles. These virus-induced structures accumulate in large clusters in the perinuclear region and primarily contain double-membrane vesicles (DMVs). See Ogando et al., “SARS-Coronavirus-2 Replication in Vero E6 Cells: Replication Kinetics, Rapid Adaptation, and Cytopathology.” 2020. J. Gen. Virol. doi: 10.1099/jgv.0.001453 PMID 32568027. Courtesy of Ronald Limpens, Montse Bárcena and Eric Snijder, Leiden University Medical Center, the Netherlands.
Transmission electron micrograph of High Five cells infected with Autographa californica multiple nucleopolyhedrovirus (AcMNPV). Image shows a large vesicle containing more than one enveloped nucleocapsid. Courtesy of Xiao-Wen Cheng, Miami University.
“Beauty Is Infectious.” Immunofluorescence image of a virus plaque formed by human metapneumovirus (HMPV) in LLC-MK2 cells. HMPV (red) and nucleus (DAPI, blue). Image courtesy of Jiuyang Xu (visting scholar, Tsinghua University School of Medicine) and John V. Williams, Department of Pediatrics, University of Pittsburgh.
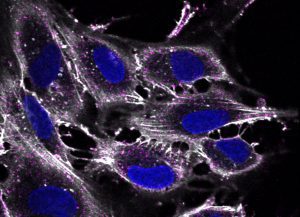
Epithelial cell monolayer (ARPE-19) infected with human cytomegalovirus, costained for viral immediate-early proteins 1/2 (green) and viral single-stranded DNA binding protein UL57 (red). Nuclei are stained with Hoechst 33342 (blue). See Vo, M., A. Aguiar, M.A. McVoy, and L. Hertel. 2020. Cytomegalovirus Strain TB40/E Restrictions and Adaptations to Growth in ARPE-19 Epithelial Cells. Microorganisms 8:E615. doi: 10.3390/microorganisms8040615. Courtesy of Alexis Aguiar and Laura Hertel, UC San Francisco.
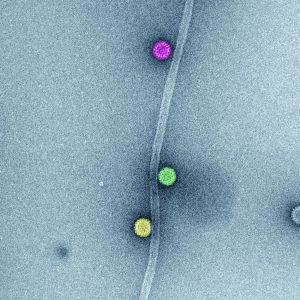
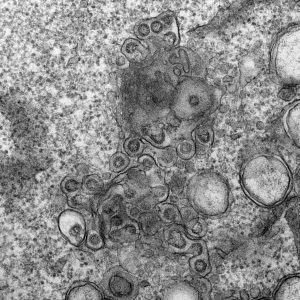
Rotavirus triple-layered particles. Rotavirus virions are triple-layered particles with icosahedral symmetry. The spike protein and the glycoprotein VP7 compose the external layer. Meanwhile, a unique component, VP6, is part of the middle layer. The VP2 icosahedral internal layer or virus core encapsidates the eleven dsRNA genome segments. All twelve vertices of inner core contain VP1-VP3 complexes. The image represents high-definition transmission electron microscopy of rotavirus triple-layered particles purified from cell culture. After glutaraldehyde fixation, the particles were stained phosphotungstate. Acquired images were colored using Adobe Photoshop. Courtesy of Catherine Eichwald, Institute of Virology, University of Zürich.
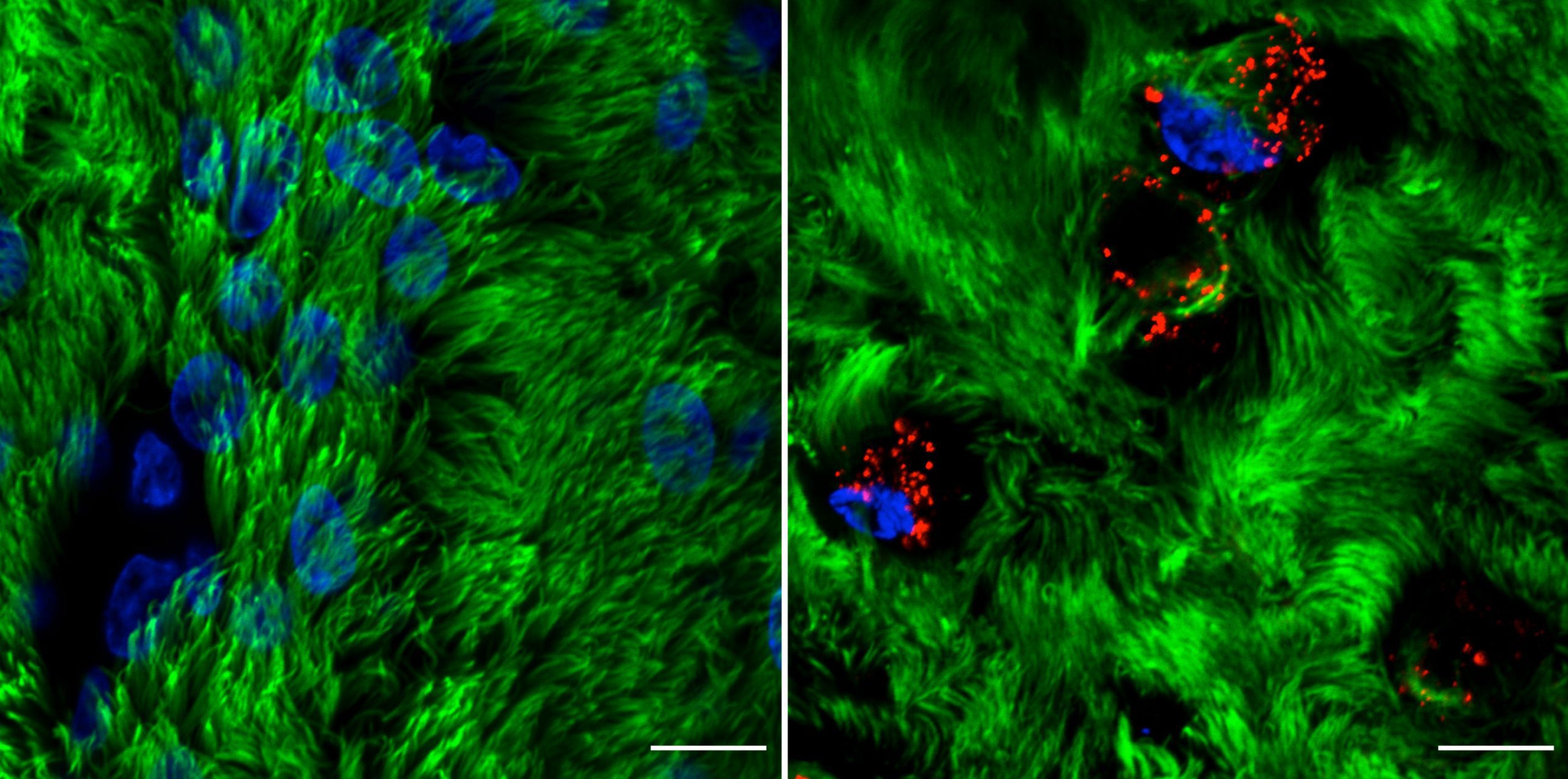
Rhinovirus C15 (RV-C15) infection of well-differentiated human bronchial epithelial cells cultured in air-liquid interface (hBEC-ALI) fixed at 12h post-infection (p.i.). A total of 50 μL of inoculum (PBS, left; or 1012 copies of RNA of RV-C15, right) was added onto the top of hBEC and incubated at 34C. The inoculum was removed 4 h p.i., and the cells were fixed at 12h p.i. with 4% paraformaldehyde. The indirect immunofluorescence assay was done using antibody against acetylated alpha-tubulin to detect motile cilia (#ab24610, Abcam; green-colored) and against dsRNA to detect the replicative intermediary forms of RV-C15 (#10010200, Scicons; red-colored); while the nuclei were stained with Hoechst 33342 (#H3570, Molecular Probes; blue-colored). The images were acquired using confocal microscopy (LSM710, Carl Zeiss Microscopy) at 630x magnification (scale bar = 10 μM). Courtesy of Talita Gagliardi, University of Maryland, College Park.
Electron micrograph of Vero E6 cells infected with SARS-CoV-2, 10 h post infection. Virus particles budding into membranes of the ERGIC. See Ogando et al., “SARS-Coronavirus-2 Replication in Vero E6 Cells: Replication Kinetics, Rapid Adaptation, and Cytopathology.” 2020. J. Gen. Virol. doi: 10.1099/jgv.0.001453 PMID 32568027. Courtesy of Ronald Limpens, Montse Bárcena and Eric Snijder, Leiden University Medical Center, the Netherlands.
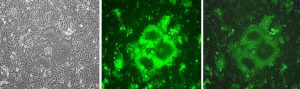
Adaptation of canine distemper virus (CDV) to human SLAM (CD150). Wild-type CDV 5804P-eGFP/H strain adapts to grow in human SLAM-expressing Vero cells (Vero-hSLAM) within five blind-passages and reproduces a skull-like syncytium, suggesting that CDV may be a potential pathogen of humans in future! Left, phase-contrast image of infected Vero-hSLAM; center, eGFP fluorescence; right, merge. Background: Expansions of the CDV host range have taken place several times during the last decades. For example, infections of monkeys (Macaca fuscata and Macaca mulata) with CDV, resulting in quite high case fatality rates, have been observed. These results suggest that CDV may eventually infect humans and spread in the human population. We investigated the requirements of wild-type CDV adaptation to the cognate human cellular measles virus SLAM receptor. This adaptation process swiftly occurred in only five virus passages. We found that adaptation of CDV to hSLAM occurred with only one amino acid exchange in the viral H protein (Manuscript in preparation). Courtesy of Jianjun Zhao1,2 & Chris. D. Richardson2 1. College of Animal Science and Veterinary Medicine, Heilongjiang Bayi Agricultural University, Daqing 163319, China; E-mail: [email protected] 2. The Department of Microbiology and Immunology, Dalhousie University, Halifax, B3H1X5, NS, Canada; E-mail: [email protected]
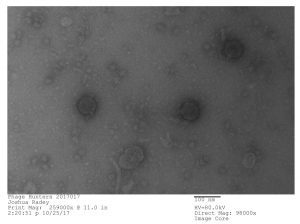
Bacteriophage discovered by Joshua Radey, in the SEA-PHAGES program, 2017. Microbacterium phage “Tiny Timothy” (Podoviridae). Courtesy of Joshua Radey and Maria D. Gainey, Western Carolina University.
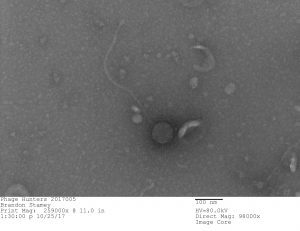
Bacteriophage discovered by Brandon Stamey, in the SEA-PHAGES program, 2017. Gordonia bacteriophage “Culver” (Siphoviridae). Courtesy of Brandon Stamey and Maria D. Gainey, Western Carolina University.
Electron micrograph of Vero E6 cells infected with SARS-CoV-2, 10 h post infection. Overview of an infected cell showing replication organelles (top right) and virus particles released from and associated with the plasma membrane. See Ogando et al., “SARS-Coronavirus-2 Replication in Vero E6 Cells: Replication Kinetics, Rapid Adaptation, and Cytopathology.” 2020. J. Gen. Virol. doi: 10.1099/jgv.0.001453 PMID 32568027. Courtesy of Ronald Limpens, Montse Bárcena and Eric Snijder, Leiden University Medical Center, the Netherlands.
Electron micrograph of Vero E6 cells infected with SARS-CoV-2, 10 h post infection. Virus particles released from and associated with the plasma membrane. See Ogando et al., “SARS-Coronavirus-2 Replication in Vero E6 Cells: Replication Kinetics, Rapid Adaptation, and Cytopathology.” 2020. J. Gen. Virol. doi: 10.1099/jgv.0.001453 PMID 32568027. Courtesy of Ronald Limpens, Montse Bárcena and Eric Snijder, Leiden University Medical Center, the Netherlands.
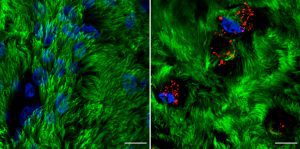
Reovirus in vivo interaction platform. The C-terminus of mammalian orthoreovirus protein μNS (amino acid region 471-721) forms cytosolic inclusions similar to globular mammalian orthoreovirus viral factories. These inclusions are directly detected by fusion to fluorescent proteins, such as mCherry, allowing their use as in vivo interaction platforms when fused to a protein of interest. For example, rotavirus NSP2, a described microtubule (MT) interactor, fused to mCherry-(471-721)μNS, allowed the localization of MTs in the interaction platforms. The image represents confocal immunofluorescence microscopy at 24 h post-transfection of CV-1 cells expressing NSP2-mCherry-(471-721)μNS (red) followed by immunostaining of microtubules (anti-tubulin, green). The NSP2-MT interaction is visualized in yellow. Nuclei were stained with DAPI (blue). Courtesy of Catherine Eichwald, Institute of Virology, University of Zürich.
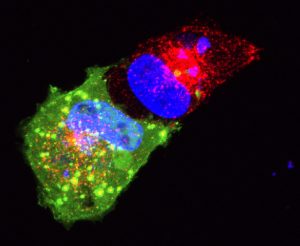
Nucleus of herpes simplex virus type 1-infected MRC-5 cell at 4 h post infection. Rendered Z-stack image includes DAPI (blue), original infecting viral DNA from the virion (green), and ICP4 associating with infecting and replicated viral DNA (red). Courtesy of Jill Dembowski, Department of Biological Sciences, Duquesne University.